貨號(hào)
產(chǎn)品規(guī)格
售價(jià)
備注
BN40459R-100ul
100ul
¥2360.00
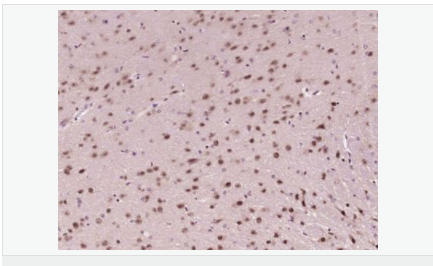
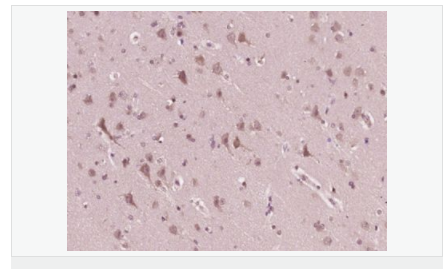
交叉反應(yīng):Human,Mouse(predicted:Rat,Dog,Pig,Rabbit) 推薦應(yīng)用:WB,IHC-P,IHC-F,ICC,IF,ELISA
BN40459R-200ul
200ul
¥3490.00
交叉反應(yīng):Human,Mouse(predicted:Rat,Dog,Pig,Rabbit) 推薦應(yīng)用:WB,IHC-P,IHC-F,ICC,IF,ELISA
產(chǎn)品描述
| 英文名稱 | RAD23B |
| 中文名稱 | 紫外切除修復(fù)蛋白R(shí)AD23B抗體 |
| 別 名 | hHR 23b; hHR23B; HR 23B; HR23B; mHR 23B; mHR23B; P58; RAD 23B; RAD23 (S. cerevisiae) homolog B; RAD23 homolog B (S. cerevisiae); RAD23 homolog B; RAD23 yeast homolog of B; RD23B_HUMAN; UV excision repair protein RAD23 homolog B; XP C repair complementing complex 58 kDa; XP C repair complementing complex 58 kDa protein; XP C repair complementing protein; XP-C repair-complementing complex 58 kDa protein; XPC repair complementing complex 58 kDa; XPC repair complementing complex 58 kDa protein; XPC repair complementing protein. |
| 研究領(lǐng)域 | 染色質(zhì)和核信號(hào) 表觀遺傳學(xué) |
| 抗體來(lái)源 | Rabbit |
| 克隆類型 | Polyclonal |
| 交叉反應(yīng) | Human, Mouse, (predicted: Rat, Dog, Pig, Rabbit, ) |
| 產(chǎn)品應(yīng)用 | WB=1:500-2000 ELISA=1:5000-10000 IHC-P=1:100-500 IHC-F=1:100-500 ICC=1:100-500 IF=1:100-500 (石蠟切片需做抗原修復(fù)) not yet tested in other applications. optimal dilutions/concentrations should be determined by the end user. |
| 分 子 量 | 43kDa |
| 細(xì)胞定位 | 細(xì)胞核 細(xì)胞漿 |
| 性 狀 | Liquid |
| 濃 度 | 1mg/ml |
| 免 疫 原 | KLH conjugated synthetic peptide derived from human hHR23b:24-120/409 |
| 亞 型 | IgG |
| 純化方法 | affinity purified by Protein A |
| 儲(chǔ) 存 液 | 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. |
| 保存條件 | Shipped at 4℃. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles. |
| PubMed | PubMed |
| 產(chǎn)品介紹 | The protein encoded by this gene is one of two human homologs of Saccharomyces cerevisiae Rad23, a protein involved in the nucleotide excision repair (NER). This protein was found to be a component of the protein complex that specifically complements the NER defect of xeroderma pigmentosum group C (XP-c) cell extracts in vitro. This protein was also shown to interact with, and elevate the nucleotide excision activity of 3-methyladenine-DNA glycosylase (MPG), which suggested a role in DNA damage recognition in base excision repair. This protein contains an N-terminal ubiquitin-like domain, which was reported to interact with 26S proteasome, and thus this protein may be involved in the ubiquitin mediated proteolytic pathway in cells. Alternative splicing results in multiple transcript variants encoding distinct isoforms. [provided by RefSeq, Sep 2011] Function: Multiubiquitin chain receptor involved in modulation of proteasomal degradation. Binds to polyubiquitin chains. Proposed to be capable to bind simultaneously to the 26S proteasome and to polyubiquitinated substrates and to deliver ubiquitinated proteins to the proteasome. May play a role in endoplasmic reticulum-associated degradation (ERAD) of misfolded glycoproteins by association with PNGase and delivering deglycosylated proteins to the proteasome. Involved in global genome nucleotide excision repair (GG-NER) by acting as component of the XPC complex. Cooperatively with CETN2 appears to stabilize XPC. May protect XPC from proteasomal degradation. The XPC complex is proposed to represent the first factor bound at the sites of DNA damage and together with other core recognition factors, XPA, RPA and the TFIIH complex, is part of the pre-incision (or initial recognition) complex. The XPC complex recognizes a wide spectrum of damaged DNA characterized by distortions of the DNA helix such as single-stranded loops, mismatched bubbles or single stranded overhangs. The orientation of XPC complex binding appears to be crucial for inducing a productive NER. XPC complex is proposed to recognize and to interact with unpaired bases on the undamaged DNA strand which is followed by recruitment of the TFIIH complex and subsequent scanning for lesions in the opposite strand in a 5'-to-3' direction by the NER machinery. Cyclobutane pyrimidine dimers (CPDs) which are formed upon UV-induced DNA damage esacpe detection by the XPC complex due to a low degree of structural perurbation. Instead they are detected by the UV-DDB complex which in turn recruits and cooperates with the XPC complex in the respective DNA repair. In vitro, the XPC:RAD23B dimer is sufficient to initiate NER; it preferentially binds to cisplatin and UV-damaged double-stranded DNA and also binds to a variety of chemically and structurally diverse DNA adducts. XPC:RAD23B contacts DNA both 5' and 3' of a cisplatin lesion with a preference for the 5' side. XPC:RAD23B induces a bend in DNA upon binding. XPC:RAD23B stimulates the activity of DNA glycosylases TDG and SMUG1. Subunit: Component of the XPC complex composed of XPC, RAD23B and CETN2. Interacts with NGLY1 and PSMC1. Interacts with ATXN3. Interacts with PSMD4 and PSMC5. Interacts with AMFR. Interacts with VCP; the interaction is indirect and mediated by NGLY1. Subcellular Location: Nucleus. Cytoplasm. Note=The intracellular distribution is cell cycle dependent. Localized to the nucleus and the cytoplasm during G1 phase. Nuclear levels decrease during S-phase; upon entering mitosis, relocalizes in the cytoplasm without association with chromatin. Similarity: Belongs to the RAD23 family. Contains 1 STI1 domain. Contains 2 UBA domains. Contains 1 ubiquitin-like domain. SWISS: P54727 Gene ID: 5887 Database links: Entrez Gene: 5887 Human Entrez Gene: 19359 Mouse Omim: 600062 Human SwissProt: P54727 Human SwissProt: P54728 Mouse Unigene: 521640 Human Unigene: 196846 Mouse Unigene: 67042 Rat Important Note: This product as supplied is intended for research use only, not for use in human, therapeutic or diagnostic applications. |