貨號
產(chǎn)品規(guī)格
售價(jià)
備注
BN41345R-100ul
100ul
¥2360.00
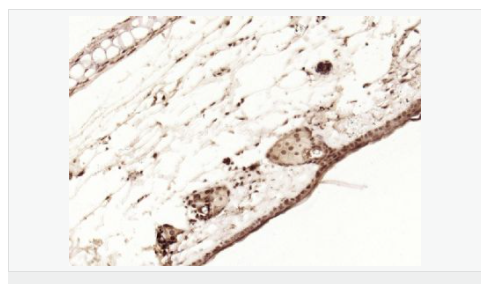
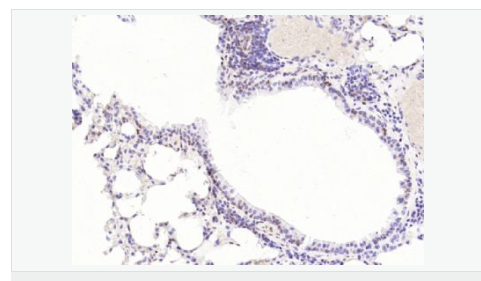
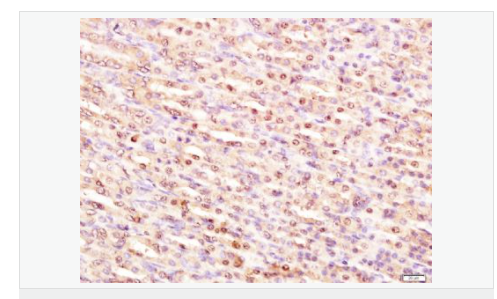
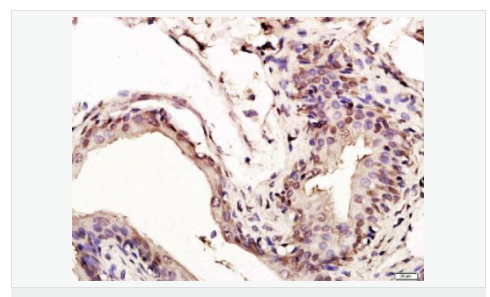
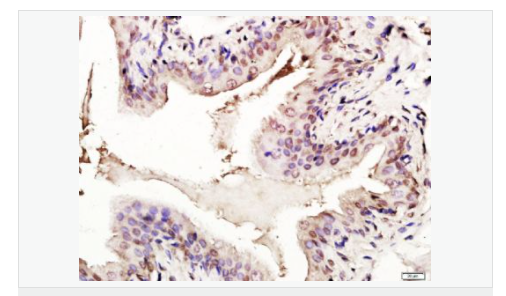
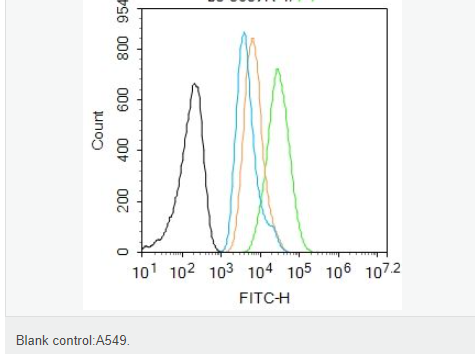
交叉反應(yīng):Human,Mouse,Rat(predicted:Dog,Pig,Cow,Horse,Sheep) 推薦應(yīng)用:WB,IHC-P,IHC-F,ICC,IF,Flow-Cyt,ELISA
BN41345R-200ul
200ul
¥3490.00
交叉反應(yīng):Human,Mouse,Rat(predicted:Dog,Pig,Cow,Horse,Sheep) 推薦應(yīng)用:WB,IHC-P,IHC-F,ICC,IF,Flow-Cyt,ELISA
產(chǎn)品描述
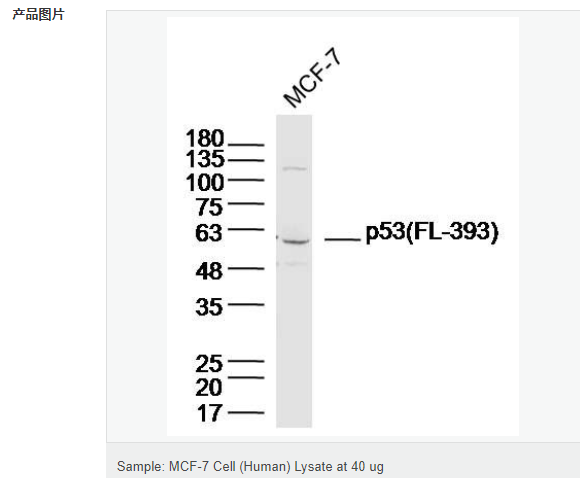
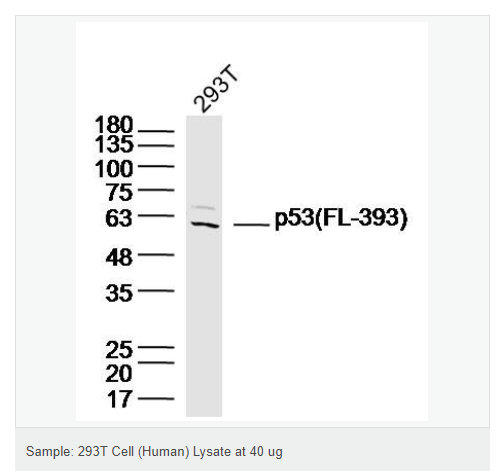
| 英文名稱 | p53 (FL-393) |
| 中文名稱 | 腫瘤抑制基因p53抗體 |
| 別 名 | LFS1; p53; p53 Cellular Tumor Antigen; p53 Tumor Suppressor; Phosphoprotein p53; TP53; Transformation related protein 53; TRP53; Tumor protein p53; Tumour Protein p53; Cellular tumor antigen p53. |
| 抗體來源 | Rabbit |
| 克隆類型 | Polyclonal |
| 交叉反應(yīng) | Human, Mouse, Rat, (predicted: Dog, Pig, Cow, Horse, Sheep, ) |
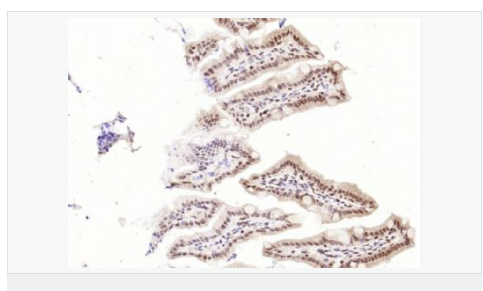
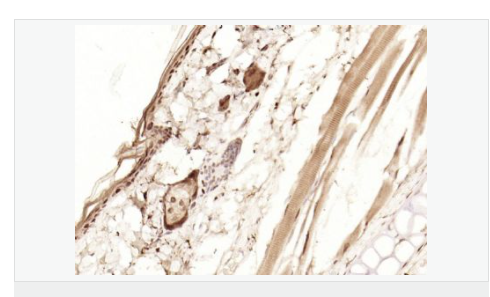
| 產(chǎn)品應(yīng)用 | WB=1:1000-2000 ELISA=1:1000-5000 IHC-P=1:100-500 IHC-F=1:100-500 Flow-Cyt=1ug/Test ICC=1:100-500 IF=1:100-500 (石蠟切片需做抗原修復(fù)) not yet tested in other applications. optimal dilutions/concentrations should be determined by the end user. |
| 分 子 量 | 53kDa |
| 細(xì)胞定位 | 細(xì)胞核 細(xì)胞漿 |
| 性 狀 | Liquid |
| 濃 度 | 1mg/ml |
| 免 疫 原 | Full length p53 protein of human origin:1-393/393 |
| 亞 型 | IgG |
| 純化方法 | affinity purified by Protein A |
| 儲 存 液 | 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. |
| 保存條件 | Shipped at 4℃. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles. |
| PubMed | PubMed |
| 產(chǎn)品介紹 | p53, a DNA-binding, oligomerization domain- and transcription activation domain-containing tumor suppressor, upregulates growth arrest and apoptosis-related genes in response to stress signals, thereby influencing programmed cell death, cell differentiation, and cell cycle control mechanisms. p53 localizes to the nucleus, yet can be chaperoned to the cytoplasm by the negative regulator, MDM2. MDM2 is an E3 ubiquitin ligase that is upregulated in the presence of active p53, where it poly-ubiquitinates p53 for proteasome targeting. p53 fluctuates between latent and active DNA-binding conformations and is differentially activated through posttranslational modifications, including phosphorylation and acetylation. Mutations in the DNA-binding domain (DBD) of p53, amino acids 110-286, can compromise energetically-favorable association with cis elements and are implicated in several human cancers. Function: [FUNCTION] Acts as a tumor suppressor in many tumor types; induces growth arrest or apoptosis depending on the physiological circumstances and cell type. Involved in cell cycle regulation as a trans-activator that acts to negatively regulate cell division by controlling a set of genes required for this process. One of the activated genes is an inhibitor of cyclin-dependent kinases. Apoptosis induction seems to be mediated either by stimulation of BAX and FAS antigen expression, or by repression of Bcl-2 expression. Implicated in Notch signaling cross-over. Prevents CDK7 kinase activity when associated to CAK complex in response to DNA damage, thus stopping cell cycle progression. Isoform 2 enhances the transactivation activity of isoform 1 from some but not all TP53-inducible promoters. Isoform 4 suppresses transactivation activity and impairs growth suppression mediated by isoform 1. Isoform 7 inhibits isoform 1-mediated apoptosis. Subunit: Interacts with AXIN1. Probably part of a complex consisting of TP53, HIPK2 and AXIN1 (By similarity). Binds DNA as a homotetramer. Interacts with histone acetyltransferases EP300 and methyltransferases HRMT1L2 and CARM1, and recruits them to promoters Subcellular Location: Cytoplasm. Nucleus. Nucleus, PML body. Endoplasmic reticulum. Note=Interaction with BANP promotes nuclear localization. Recruited into PML bodies together with CHEK2. Tissue Specificity: Ubiquitous. Isoforms are expressed in a wide range of normal tissues but in a tissue-dependent manner. Isoform 2 is expressed in most normal tissues but is not detected in brain, lung, prostate, muscle, fetal brain, spinal cord and fetal liver. Isoform 3 is expressed in most normal tissues but is not detected in lung, spleen, testis, fetal brain, spinal cord and fetal liver. Isoform 7 is expressed in most normal tissues but is not detected in prostate, uterus, skeletal muscle and breast. Isoform 8 is detected only in colon, bone marrow, testis, fetal brain and intestine. Isoform 9 is expressed in most normal tissues but is not detected in brain, heart, lung, fetal liver, salivary gland, breast or intestine. Post-translational modifications: Acetylated. Acetylation of Lys-382 by CREBBP enhances transcriptional activity. Deacetylation of Lys-382 by SIRT1 impairs its ability to induce proapoptotic program and modulate cell senescence. Phosphorylation on Ser residues mediates transcriptional activation. Phosphorylated by HIPK1. Phosphorylation at Ser-9 by HIPK4 increases repression activity on BIRC5 promoter. Phosphorylated on Thr-18 by VRK1. Phosphorylated on Ser-20 by CHEK2 in response to DNA damage, which prevents ubiquitination by MDM2. Phosphorylated on Ser-20 by PLK3 in response to reactive oxygen species (ROS), promoting p53/TP53-mediated apoptosis. Phosphorylated on Thr-55 by TAF1, which promotes MDM2-mediated degradation. Phosphorylated on Ser-33 by CDK7 in a CAK complex in response to DNA damage. Phosphorylated on Ser-46 by HIPK2 upon UV irradiation. Phosphorylation on Ser-46 is required for acetylation by CREBBP. Phosphorylated on Ser-392 following UV but not gamma irradiation. Phosphorylated upon DNA damage, probably by ATM or ATR. Phosphorylated on Ser-15 upon ultraviolet irradiation; which is enhanced by interaction with BANP. Phosphorylated by NUAK1 at Ser-15 and Ser-392; was initially thought to be mediated by STK11/LKB1 but it was later shown that it is indirect and that STK11/LKB1-dependent phosphorylation is probably mediated by downstream NUAK1 (PubMed:21317932). It is unclear whether AMP directly mediates phosphorylation at Ser-15. Phosphorylated on Thr-18 by isoform 1 and isoform 2 of VRK2. Phosphorylation on Thr-18 by isoform 2 of VRK2 results in a reduction in ubiquitination by MDM2 and an increase in acetylation by EP300. Stabilized by CDK5-mediated phosphorylation in response to genotoxic and oxidative stresses at Ser-15, Ser-33 and Ser-46, leading to accumulation of p53/TP53, particularly in the nucleus, thus inducing the transactivation of p53/TP53 target genes. Phosphorylated at Ser-315 and Ser-392 by CDK2 in response to DNA-damage. Dephosphorylated by PP2A-PPP2R5C holoenzyme at Thr-55. SV40 small T antigen inhibits the dephosphorylation by the AC form of PP2A. May be O-glycosylated in the C-terminal basic region. Studied in EB-1 cell line. Ubiquitinated by MDM2 and SYVN1, which leads to proteasomal degradation. Ubiquitinated by RFWD3, which works in cooperation with MDM2 and may catalyze the formation of short polyubiquitin chains on p53/TP53 that are not targeted to the proteasome. Ubiquitinated by MKRN1 at Lys-291 and Lys-292, which leads to proteasomal degradation. Deubiquitinated by USP10, leading to its stabilization. Ubiquitinated by TRIM24, which leads to proteasomal degradation. Ubiquitination by TOPORS induces degradation. Deubiquitination by USP7, leading to stabilization. Isoform 4 is monoubiquitinated in an MDM2-independent manner. Monomethylated at Lys-372 by SETD7, leading to stabilization and increased transcriptional activation. Monomethylated at Lys-370 by SMYD2, leading to decreased DNA-binding activity and subsequent transcriptional regulation activity. Lys-372 monomethylation prevents interaction with SMYD2 and subsequent monomethylation at Lys-370. Dimethylated at Lys-373 by EHMT1 and EHMT2. Monomethylated at Lys-382 by SETD8, promoting interaction with L3MBTL1 and leading to repress transcriptional activity. Demethylation of dimethylated Lys-370 by KDM1A prevents interaction with TP53BP1 and represses TP53-mediated transcriptional activation. Sumoylated by SUMO1. DISEASE: Note=TP53 is found in increased amounts in a wide variety of transformed cells. TP53 is frequently mutated or inactivated in about 60% of cancers. TP53 defects are found in Barrett metaplasia a condition in which the normally stratified squamous epithelium of the lower esophagus is replaced by a metaplastic columnar epithelium. The condition develops as a complication in approximately 10% of patients with chronic gastroesophageal reflux disease and predisposes to the development of esophageal adenocarcinoma. Similarity: Belongs to the p53 family. SWISS: P04637 Gene ID: 7157 Database links: Entrez Gene: 7157 Human Entrez Gene: 22059 Mouse Omim: 191170 Human SwissProt: P04637 Human SwissProt: P02340 Mouse Unigene: 654481 Human Unigene: 222 Mouse Unigene: 54443 Rat Important Note: This product as supplied is intended for research use only, not for use in human, therapeutic or diagnostic applications. |